New developments in genetics and metagenomics over the past 15 years have led scientists to produce an in-depth characterization of the composition and function of the gut microbiome as a novel organ in the close intersection between health and disease. As a result, the number of publications discussing the gut microbiota over the past five years represents more than 80% of all publications over the past 40 years on the topic, thus reflecting how microbiome research is changing basic science and medicine.
The human gut microbiome has been linked to the risk and even the onset of clinical symptoms of not only gut-related diseases, but also disorders that apparently have no direct relationship with the gut, such as cardiometabolic disorders, neuropsychiatric diseases and cancer. As a result, the human gut microbiota is viewed as a potential source of new therapeutics. However, the role of bacteria on specific phenotypes based on changes in their abundance does not mean we can infer that specific species have either a protective or detrimental effect on the host.
In the human body, microorganisms constantly interact with host cells at different levels and the connections established between immunity, microbes and metabolism are especially interesting. In the gastrointestinal tract, immune cells dialogue with the gut microbiota and vice versa, driving a fine-tuned crosstalk between microbes and host under the influence of several factors including genes, dietary habits, drug treatments and stool consistency and frequency.
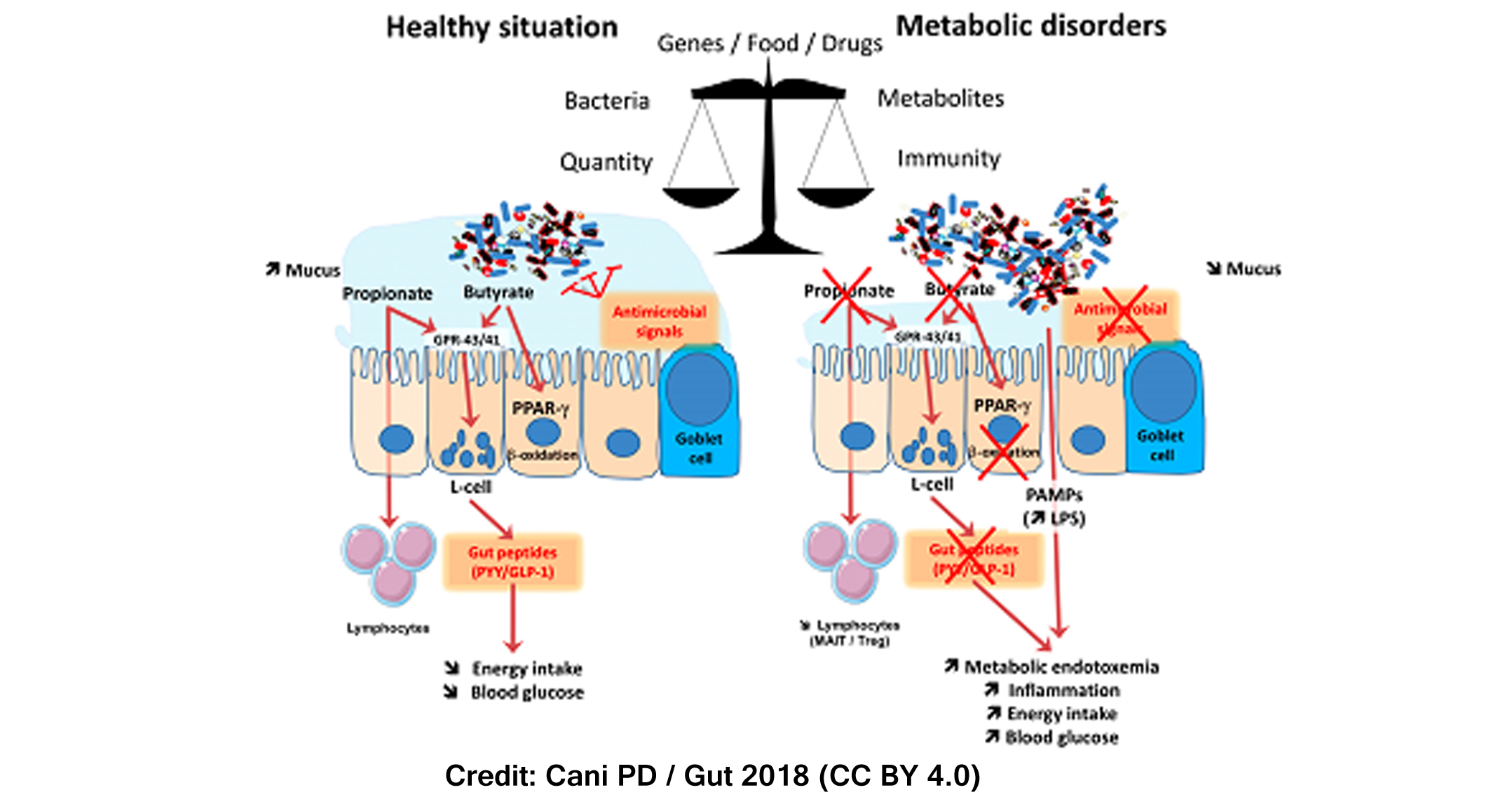
Studies carried out in our research group over the past decade have focused on the framework of the interactions between gut microbiota and host metabolism. During metabolic disorders, changes in the gut microbiome may be linked to reduced mucus thickness, decreased antimicrobial defense but also changes in butyrate and propionate production. As a consequence, the enteroendocrine L-cells found in the gut secrete less gut peptides thereby affecting several pathways ranging from the regulation of food intake (e.g., GLP-1) to maintenance of the gut barrier function (e.g., GLP-2). In addition, together with changes in the gut microbial environment and metabolites an alteration of the intestinal barrier can lead to the leakage of pathogen-associated molecular patterns (PAMPs) such as the lipopolysaccharides (LPS). Our research group demonstrated for the first time in 2007 that when LPS translocate (i.e. metabolic endotoxemia) and increases in the bloodstream, low-grade systemic inflammatory responses may be initiated, along with insulin resistance through mechanisms of interaction between gut microbes and the innate immune system.
Both the gut microbiome and the metabolites it produces can influence our metabolism. The fact that each microbial metabolite may play different roles in its relationship with the host highlights how redundancy is a feature that can be used to define a healthy gut microbiota profile. This is joined by resilience and a high abundance of health-promoting microorganisms within the indigenous ecosystem.
When studying the gut microbiome, the absolute quantity of microbes measured using quantitative microbiome profiling is preferred over the relative proportions of microbes measured with classic relative abundance-based profiling, giving a more realistic picture of what is going on. Furthermore, analyzing the metabolic capacity of the gut microbiota and the microbial metabolites provides relevant information regarding disease-specific bacterial characteristics, which is complementary to the information regarding microbial composition alone, obtained through metagenomics. These observations demonstrate the importance of multiomics approaches for better understanding the interactions between microbes, host and metabolism. However, the challenges that deserve to be covered on the path towards personalized nutritional advice based on the gut microbiome include the need for large-scale longitudinal studies focusing on both the mucosal microbiota and the luminal microbiota instead of “only” fecal material. But also using samples for studying the activity of the microbiome (e.g. blood, urine), and eventually the need for standardized protocols.
Finally, proofs of concept are required using isolated bacteria or specific metabolites in order to move away from correlations between specific microbes and diseases that do not provide the exact implication of the microbe as beneficial or deleterious. In this context, Prevotella copri and Akkermansia muciniphila are two examples of bacteria that have been largely investigated through functional proof-of-concept studies.
The beneficial or detrimental role of specific bacteria on host metabolism may be influenced by complex interactions between diet and host. This is the case of P. copri, which, in experimental studies, has been found to improve glucose metabolism and insulin sensitivity through the production of succinate from fermentable dietary fibers. However, it can also aggravate glucose intolerance and reduce insulin sensitivity when administered to mice together with a high-fat diet.
Regarding A. muciniphila, different pathological situations have been associated when the levels of this bacterium have been found to be lower (obesity and non-treated type 2 diabetes) or higher (metformin-treated type 2 diabetes, gastric bypass, multiple sclerosis and Parkinson’s disease). However, several confounding factors may modulate the gut microbiota and eventually the abundance of A. muciniphila (as for example: dietary fiber, metformin and other drug treatments as well as intestinal transit time). It is worth noting that so far, data for which a proof-of-concept of the link between the disease and the presence of the bacterium has been made are individuals with a specific condition or responders to an anti-cancer, as well as in obesity, type 2 diabetes and gastric bypass. Indeed, in our research group, we have shown that supplementation with A. muciniphila, whether alive or pasteurized, protects against several cardiometabolic features in mice and we are now working on proving whether it would be useful for achieving a long-lasting shift in humans’ microbiota.
To sum up, a comprehensive study of interactions between the gut microbiota, host and metabolism deserves a focus not only on the composition but also on the activity of these microorganisms and their metabolites. Furthermore, functional proof-of-concept studies are needed, to move away from studies comparing diseases and healthy subjects and in order to better explore the causal relationship between a given microbe and a disease or healthy situation.
Reference:
Cani PD. Human gut microbiome: hopes, threats and promises. Gut Epub ahead of print: 27/06/2018. doi: 10.1136/gutjnl-2018-316723.